New paper of Dr. Shi Yazhou published by PLoS Comput Biol!
Here is the online version!
Citation: Shi Y-Z, Jin L, Feng C-J, Tan Y-L, Tan Z-J (2018) Predicting 3D structure and stability of RNA pseudoknots in monovalent and divalent ion solutions. PLoS Comput Biol 14(6):e1006222.
https://doi.org/10.1371/journal.pcbi.1006222
Predicting 3D structure and stability of RNA pseudoknots in monovalent and divalent ion solutions

Abstract:
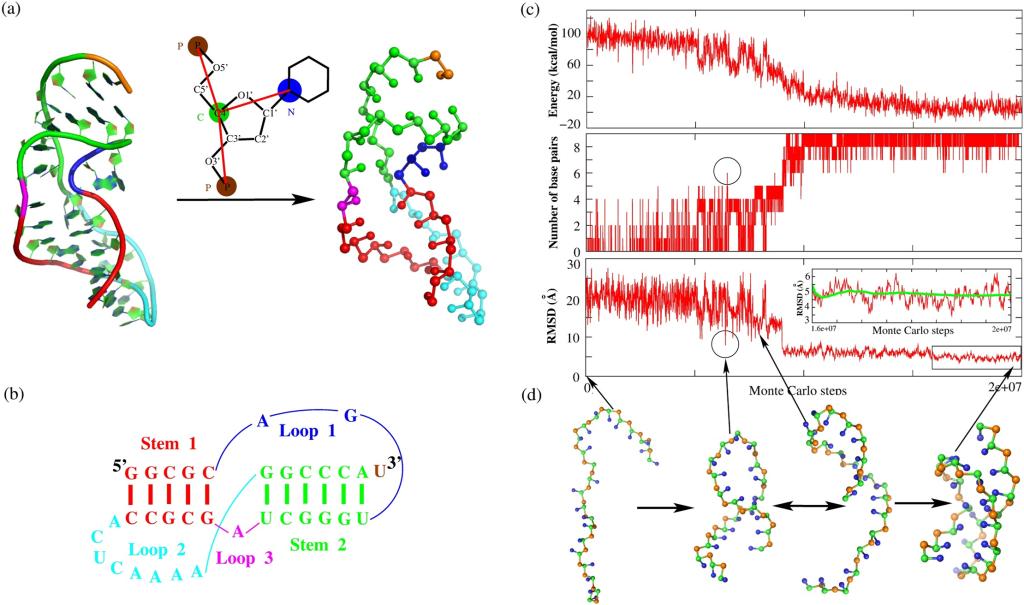
RNA pseudoknots are a kind of minimal RNA tertiary structural motifs, and their three-dimensional (3D) structures and stability play essential roles in a variety of biological functions. Therefore, to predict 3D structures and stability of RNA pseudoknots is essential for understanding their functions. In the work, we employed our previously developed coarse-grained model with implicit salt to make extensive predictions and comprehensive analyses on the 3D structures and stability for RNA pseudoknots in monovalent/divalent ion solutions. The comparisons with available experimental data show that our model can successfully predict the 3D structures of RNA pseudoknots from their sequences, and can also make reliable predictions for the stability of RNA pseudoknots with different lengths and sequences over a wide range of monovalent/divalent ion concentrations. Furthermore, we made comprehensive analyses on the unfolding pathway for various RNA pseudoknots in ion solutions. Our analyses for extensive pseudokonts and the wide range of monovalent/divalent ion concentrations verify that the unfolding pathway of RNA pseudoknots is mainly dependent on the relative stability of unfolded intermediate states, and show that the unfolding pathway of RNA pseudoknots can be significantly modulated by their sequences and solution ion conditions.